Remodeler tutorial¶
This tutorial introduces the table-remodeler tools for restructuring tabular (.tsv) files. The tools are particularly useful for transforming event files from experimental logs and reorganizing data to enable specific analyses.
The remodeling tools are written in Python and designed to operate on entire datasets. Datasets can be in BIDS format or any directory structure containing tabular files with a particular suffix (e.g., _events.tsv). The tools support multiple execution modes: command-line scripts, Python programs, Jupyter notebooks, or online tools for debugging.
This quickstart covers the core concepts of remodeling with practical examples. For comprehensive operation details, see the Operations reference and User guide.
Table of contents¶
What is remodeling?¶
Although the remodeling process can be applied to any tabular file, it is most often used for restructuring event files. Event files consist of identified time markers linked to the timeline of an experiment, providing a crucial bridge between what happens in the experiment and the experimental data.
Event files are often initially created using information in the log files generated by the experiment control software. The entries in the log files mark time points within the experimental record at which something changes or happens (such as the onset or offset of a stimulus or a participant response). These event files are then used to identify portions of the data corresponding to particular points or blocks of data to be analyzed or compared.
Remodeling refers to the process of file restructuring including creating, modifying, and reorganizing tabular files in order to disambiguate or clarify their information to enable or streamline their analysis and/or further distribution. Remodeling can occur at several stages during the acquisition and processing of experimental data as shown in this schematic diagram:
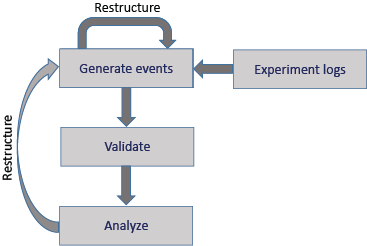
In addition to restructuring during initial structuring of the tabular files, further event file restructuring may be useful when the event files are not suited to the requirements of a particular analysis. Thus, restructuring can be an iterative process, which is supported by the table-remodeler tools for datasets with tabular event files.
Types of operations¶
Remodeling operations fall into two categories:
Transformation operations modify the tabular files by restructuring their content. They process an input pandas DataFrame and return a transformed DataFrame without modifying the original data. Transformations are stateless and include:

Clean-up: Remove/rename/reorder columns, remove rows
Factor: Extract condition variables and design matrices
Restructure: Merge consecutive events, remap values, split trial-encoded rows
Summarization operations extract information without modifying the input DataFrame. They analyze files and produce summary reports stored separately:
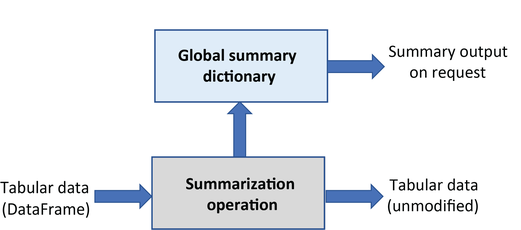
Column name and value summaries
HED tag and validation summaries
Definition summaries
Sidecar template generation
The following table summarizes all available operations:
Category |
Operation |
Example use case |
|---|---|---|
clean-up |
||
Remove temporary columns created during restructuring |
||
Remove rows with a particular value in a specified column |
||
Make columns names consistent across a dataset |
||
Make column order consistent across a dataset |
||
factor |
||
Extract factor vectors from a column of condition variables |
||
Extract factor vectors from search queries of HED annotations |
||
Extract design matrices and/or condition variables |
||
restructure |
||
Replace multiple consecutive events of the same type with one of longer duration |
||
Create m columns from values in n columns (for recoding) |
||
Split trial-encoded rows into multiple events |
||
summarization |
||
Summarize column names and order in the files |
||
Count the occurrences of the unique column values |
||
Summarize definitions used and report inconsistencies |
||
Summarize the HED tags present in the |
||
Summarize the detailed usage of a particular type tag such as Condition-variable or Task (used to automatically extract experimental designs) |
||
Validate the data files and report any errors |
||
Generate a sidecar template from an event file |
For detailed parameter descriptions and examples of each operation, see the Operations reference.
The remodeling process¶
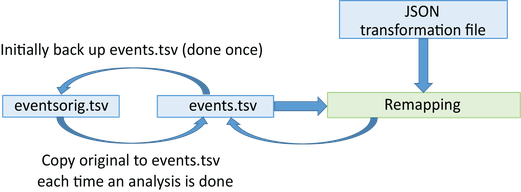
Remodeling applies a list of operations to tabular files to restructure or extract information. The following diagram shows the complete workflow:
Backup system¶
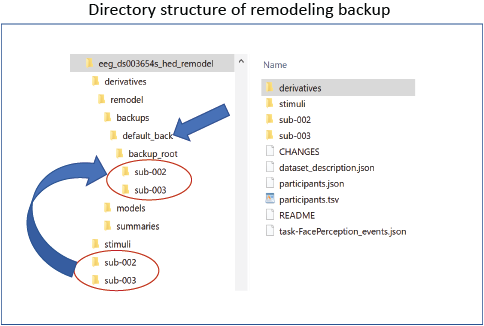
Initially, you create a backup of the tabular files you plan to remodel. This backup is performed once and stored in the derivatives/remodel/backups subdirectory of your dataset. The backup structure mirrors your original file organization:
Important: The remodeling process always reads from the backup and writes to the original file location. This means:
You can iterate on your remodeling operations without fear of losing data
Rerunning operations always starts from the original backup
You can correct mistakes in your remodeling script and rerun
Remodeling workflow¶
Create backup (one time): Use
run_remodel_backupto create the baseline backupCreate remodeling file: Write JSON file with your operations (typically named
*_rmdl.json)Run remodeling: Execute
run_remodelwhich:Reads files from backup directory
Applies operations sequentially
Writes transformed files to original locations
Saves summaries to
derivatives/remodel/summaries
Iterate: Modify your JSON file and rerun as needed
By convention, remodeling files are stored in derivatives/remodel/remodeling_files with names ending in _rmdl.json.
Checkpoints and backups¶
Advanced users can create multiple named backups to use as checkpoints:
# Create a checkpoint after initial cleanup
run_remodel_backup data_dir --backup-name "after_cleanup"
# Later, restore from that checkpoint
run_remodel_restore data_dir --backup-name "after_cleanup"
This is useful when developing different versions of remodeling for different purposes.
JSON remodeling files¶
The operations to restructure a tabular file are stored in a remodel file in JSON format. The file consists of a list of JSON dictionaries.
Basic operation syntax¶
Each operation is specified as a JSON dictionary with three components: the operation name, a description, and parameters. Here’s a simple example that renames a column:
Example of a rename operation.
{
"operation": "rename_columns",
"description": "Rename trial_type column to event_type for consistency",
"parameters": {
"column_mapping": {
"trial_type": "event_type"
},
"ignore_missing": true
}
}
Each operation has its own specific required and optional parameters. For rename_columns:
column_mapping (required): Dictionary mapping old names to new names
ignore_missing (required): If
true, don’t error when a column doesn’t exist
See the Operations reference for detailed parameter documentation.
Applying multiple operations¶
A remodeling JSON file contains a list of operations executed sequentially. Each operation sees the result of previous operations. For example, here we rename columns and then summarize the result:
Multiple operations in sequence.
[
{
"operation": "rename_columns",
"description": "Rename trial_type to event_type for consistency.",
"parameters": {
"column_mapping": {
"trial_type": "event_type"
},
"ignore_missing": true
}
},
{
"operation": "summarize_column_names",
"description": "Verify column names after renaming.",
"parameters": {
"summary_name": "Columns after remodeling",
"summary_filename": "columns_after_remodel"
}
}
]
Important: Remodeling always starts from the backup. If you run this script multiple times, each run begins with the original backup files, applies all operations in order, and overwrites the data files (but not the backup). This means you can safely iterate on your remodeling script.
Complex remodeling¶
This section demonstrates a realistic remodeling scenario using an excerpt from the AOMIC-PIOP2 dataset (ds002790 on OpenNeuro):
Excerpt from AOMIC stop-signal task event file.
onset |
duration |
trial_type |
stop_signal_delay |
response_time |
response_accuracy |
response_hand |
sex |
|---|---|---|---|---|---|---|---|
0.0776 |
0.5083 |
go |
n/a |
0.565 |
correct |
right |
female |
5.5774 |
0.5083 |
unsuccesful_stop |
0.2 |
0.49 |
correct |
right |
female |
9.5856 |
0.5084 |
go |
n/a |
0.45 |
correct |
right |
female |
13.5939 |
0.5083 |
succesful_stop |
0.2 |
n/a |
n/a |
n/a |
female |
17.1021 |
0.5083 |
unsuccesful_stop |
0.25 |
0.633 |
correct |
left |
male |
21.6103 |
0.5083 |
go |
n/a |
0.443 |
correct |
left |
male |
Understanding the task¶
This stop-signal task presented faces to participants who decided the sex by pressing a button (left/right hand). However, if a stop signal occurred, participants should refrain from responding.
Trial vs. event encoding¶
The file uses trial-level encoding: each row represents an entire trial with multiple events encoded as offsets:
The
onsetcolumn marks the face presentation (go signal)The
stop_signal_delaycolumn contains the time delay of the stop signal (if present) from the onsetThe
response_timecolumn contains the time delay of the button press (if present) from the onset
For many analyses, event-level encoding is preferable: each row represents a single event.
Applying split_rows¶
The split_rows operation converts trial encoding to event encoding:
Splitting trial-level rows into events.
[
{
"operation": "split_rows",
"description": "Convert from trial encoding to event encoding.",
"parameters": {
"anchor_column": "trial_type",
"new_events": {
"response": {
"onset_source": ["response_time"],
"duration": [0],
"copy_columns": ["response_accuracy", "response_hand"]
},
"stop_signal": {
"onset_source": ["stop_signal_delay"],
"duration": [0.5]
}
},
"remove_parent_row": false
}
}
]
Understanding the parameters¶
anchor_column: Specifies which column receives the new event type values (trial_type in this case). Since remove_parent_row is false, the original trial row remains.
new_events: Dictionary where each key is a new event type name. Each event specification has:
onset_source: List of column names and/or numbers to compute the new event’s onset. Values are added to the parent row’s onset. Column names are evaluated; if any value is
n/aor missing, no new event is created.["response_time"]→ new onset = parent onset + value in response_time column["stop_signal_delay"]→ new onset = parent onset + value in stop_signal_delay column
duration: Duration for the new event (0 for button presses, 0.5 for stop signal)
copy_columns (optional): Columns to copy from parent row to new row. For response events, we copy
response_accuracyandresponse_hand. Other columns are filled withn/a.
Remodeling results¶
After remodeling, trials with responses will have additional rows for the response event. Trials with stop signals will have additional rows for the stop signal. The tool automatically handles missing values (n/a) without creating spurious events.
You can find the complete remodeling file at: AOMIC_splitevents_rmdl.json
Remodeling file locations¶
When executing remodeling operations, provide the full path to the JSON remodeling file. However, it’s good practice to organize remodeling files within your dataset:
Recommended directory structure:
dataset_root/
├── sub-0001/
│ └── ...
├── derivatives/
│ └── remodel/
│ ├── backups/ # Created automatically by run_remodel_backup
│ │ └── default/ # Default backup name (or custom names)
│ ├── remodeling_files/ # Store your *_rmdl.json files here
│ │ ├── cleanup_rmdl.json
│ │ ├── split_trials_rmdl.json
│ │ └── summarize_rmdl.json
│ └── summaries/ # Created automatically by summarization operations
│ ├── column_names_summary.txt
│ └── column_values_summary.json
This organization keeps all remodeling-related files together and provides a clear record of transformations applied to your dataset.
Using the tools¶
The table-remodeler provides several ways to execute remodeling operations: online tools for debugging, command-line scripts for batch processing, and Python/Jupyter for programmatic control.
Online tools for debugging¶
Before running remodeling on an entire dataset, test your operations on a single file using the HED online tools.
Steps to use the online remodeler:
Navigate to the Events page
Select the Execute remodel script action
Upload your data file (
.tsv) and JSON remodel file (*_rmdl.json)Press Process
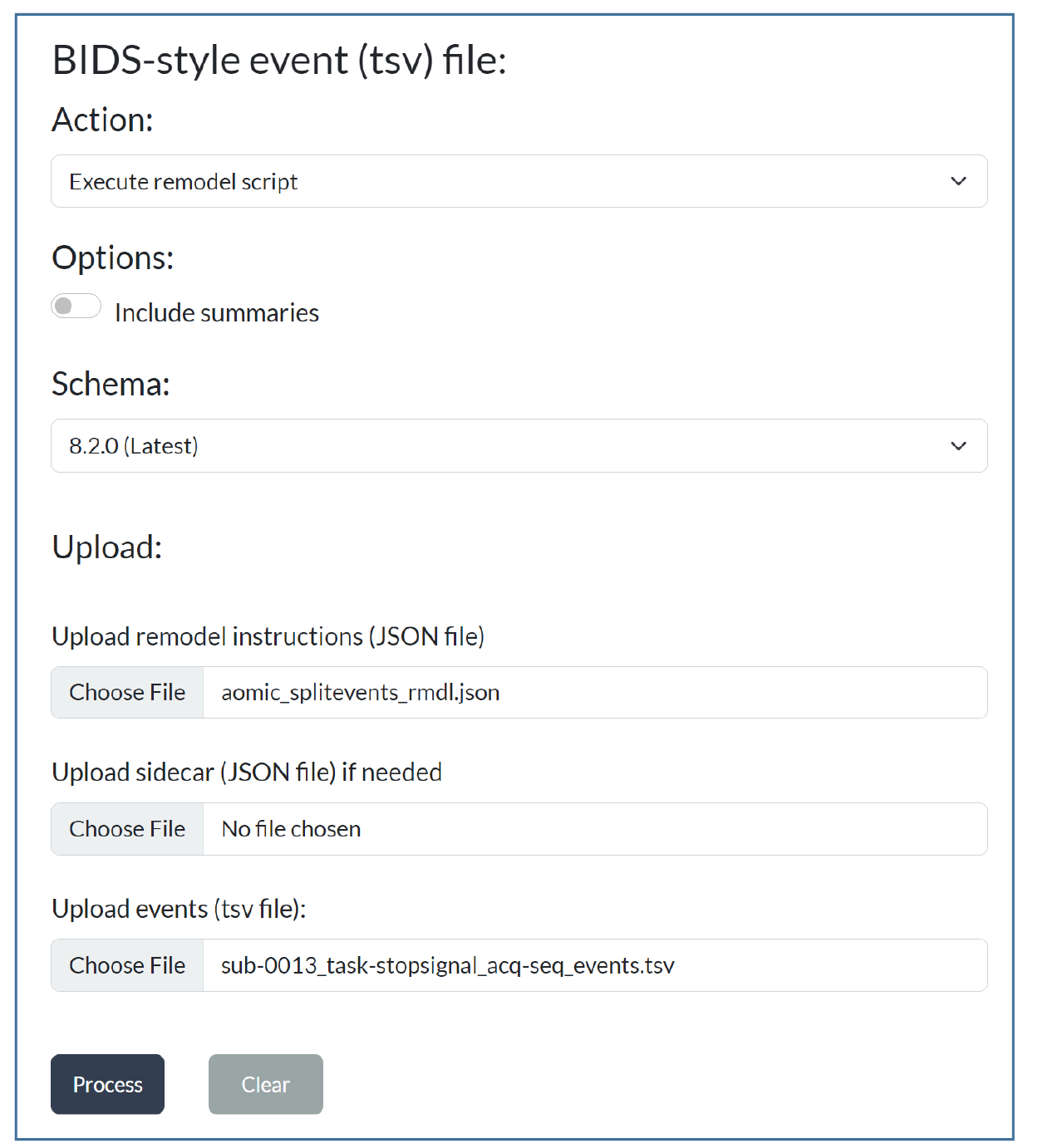
Results:
If errors exist: Downloads a text file with error descriptions
If successful (transformations only): Downloads the remodeled data file
If successful (with summaries): Downloads a zip file containing the remodeled data file and summary files
For HED operations: Also upload a JSON sidecar file containing HED annotations if using operations like factor_hed_tags, summarize_hed_validation, or other HED-dependent operations.
The online tools are ideal for:
Debugging JSON syntax errors
Verifying operation parameters
Previewing transformation results
Testing with small data samples
Command-line interface¶
After installing the table-remodeler, you can process entire datasets using the command-line scripts.
Installation¶
pip install table-remodeler
Available commands¶
Three command-line tools are provided:
run_remodel_backup: Create backup of tabular files (run once per dataset)
run_remodel: Execute remodeling operations
run_remodel_restore: Restore files from backup
Basic usage example¶
# Step 1: Create backup (one time)
run_remodel_backup /data/my_dataset
# Step 2: Run remodeling operations
run_remodel /data/my_dataset /data/my_dataset/derivatives/remodel/remodeling_files/my_operations_rmdl.json -b -x derivatives
# Step 3: Check the summaries
ls /data/my_dataset/derivatives/remodel/summaries/
Common parameters¶
The run_remodel command accepts:
data_dir (positional, required): Root directory of the dataset
model_path (positional, required): Path to JSON remodeling file
-b, –bids-format: Treat as BIDS-formatted dataset
-x, –exclude-dirs: Directories to exclude (e.g.,
derivatives)-t, –task-names: Process only files with specific task names
-s, –save-formats: Formats for summaries (
.txt,.json)-v, –verbose: Enable verbose output
AOMIC dataset example¶
run_remodel /data/ds002790 \
/data/ds002790/derivatives/remodel/remodeling_files/AOMIC_splitevents_rmdl.json \
-b -s .txt -s .json -x derivatives -t stopsignal
This command:
Processes the ds002790 dataset in BIDS format (
-b)Applies operations from
AOMIC_splitevents_rmdl.jsonSaves summaries in both text and JSON formats (
-s .txt -s .json)Excludes the derivatives directory (
-x derivatives)Only processes files with
stopsignalin the filename (-t stopsignal)
Important notes:
Always run from backup (default behavior)
Rerunning
run_remodelstarts fresh from backup each timeSummaries are saved to
derivatives/remodel/summaries/Original data files are overwritten (but backups remain safe)
For comprehensive command-line documentation, see the User guide.
Jupyter notebooks¶
For programmatic control with documentation, use the command-line scripts from within Jupyter notebooks. Example notebooks are available at table-remodeler/examples.
These notebooks demonstrate how to:
Create backups programmatically
Execute remodeling operations with custom parameters
Process summaries and visualize results
Integrate remodeling into analysis pipelines
If you don’t have access to a Jupyter environment, see Six easy ways to run your Jupyter Notebook in the cloud for no-cost cloud options.
Next steps¶
Now that you understand the basics of remodeling:
Explore operations: See the Operations reference for detailed parameter documentation
Learn advanced workflows: Read the User guide for CLI details, HED integration, and advanced topics
Create custom operations: See the Creating custom operations section of the User guide to extend Table Remodeler
Try examples: Download example datasets and remodeling files from hed-examples